After the LettuceGDB project initialized, much discussion was taken to design the basic frame of this unique database. In brief, it includes three main parts:
1) what kind of data will be included;
2) How to present these mega data;
3) How the database can benefit users/researchers.
In terms of a month discussion, we came to the frame as follows.
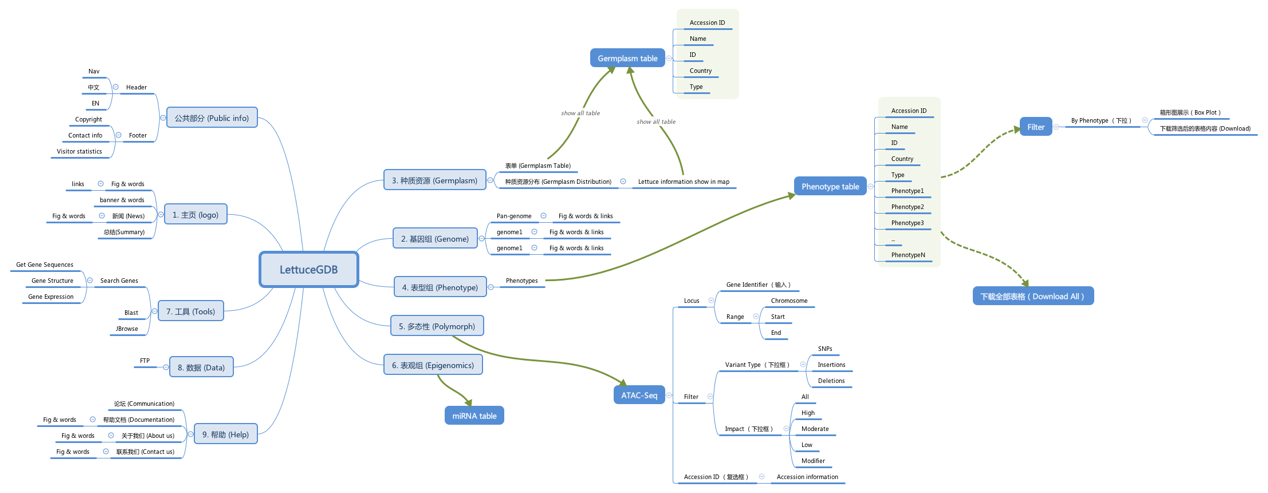
Considering the multi-omics data, including genome references, RNA-Seq datasets, sRNA-Seq datasets, PARE-Seq datasets, and ATAC-Seq dataset, etc., all together compose a huge pool of mega data. How to present these data is the key. We designed three ports to resolve this issue.
First, a flattening system will be employed to display the mega data, which means a specific interface for each type of omics data will be constructed. Specifically, LettuceGDB will include four modules, namely Genome, Germplasm, Genotype and multi-omics.
Second, we will develop a suite of tools for assisting sequence retrieval and functional exploration.
Third, to users, how to fetch/download the data they are interested. We designed an easy access by which users could download all data displayed/stored in webpages. Meanwhile, accesses for bulk and customized download ports will be deployed.
The following is an overall design of LettuceGDB by software XMind.